Data correlations evaluation module
Source:R/data_plots.R, R/plot_hbar.R, R/plot_ridge.R, and 3 more
data-plots.RdData correlations evaluation module
Wrapper to create plot based on provided type
Nice horizontal stacked bars (Grotta bars)
Plot nice ridge plot
Readying data for sankey plot
Beautiful sankey plot with option to split by a tertiary group
Beautiful violin plot
Beatiful violin plot
Usage
data_visuals_ui(id, tab_title = "Plots", ...)
data_visuals_server(id, data, ...)
create_plot(data, type, x, y, z = NULL, ...)
plot_hbars(data, x, y, z = NULL)
plot_ridge(data, x, y, z = NULL, ...)
sankey_ready(data, x, y, numbers = "count", ...)
plot_sankey(data, x, y, z = NULL, color.group = "x", colors = NULL)
plot_scatter(data, x, y, z = NULL)
plot_violin(data, x, y, z = NULL)Value
Shiny ui module
shiny server module
ggplot2 object
ggplot2 object
ggplot2 object
data.frame
ggplot2 object
ggplot2 object
ggplot2 object
Examples
create_plot(mtcars, "plot_violin", "mpg", "cyl")
#> Error in if (!z %in% names(data)) { z <- NULL}: argument is of length zero
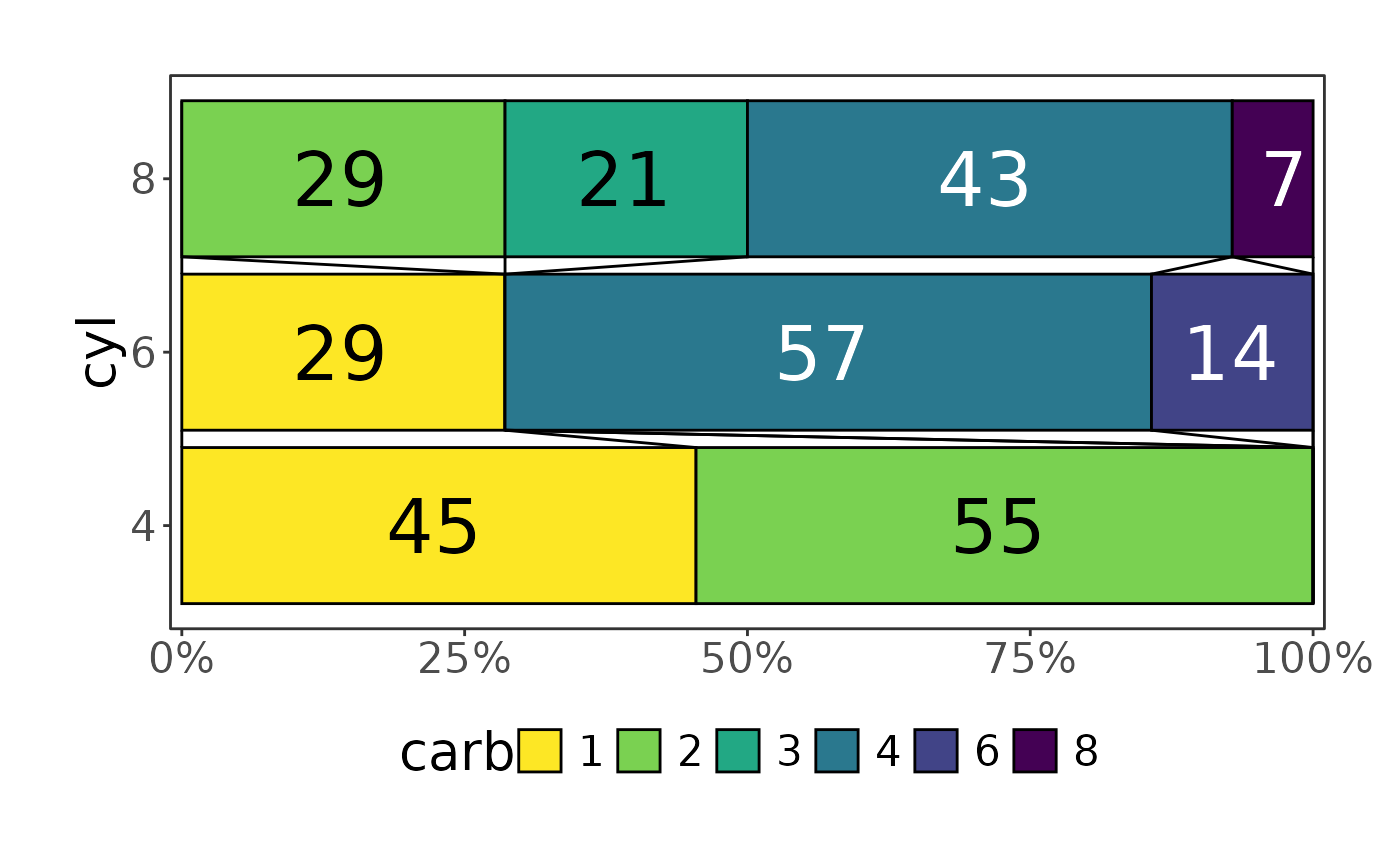
mtcars |> plot_hbars(x = "carb", y = "cyl")
#> Scale for fill is already present.
#> Adding another scale for fill, which will replace the existing scale.
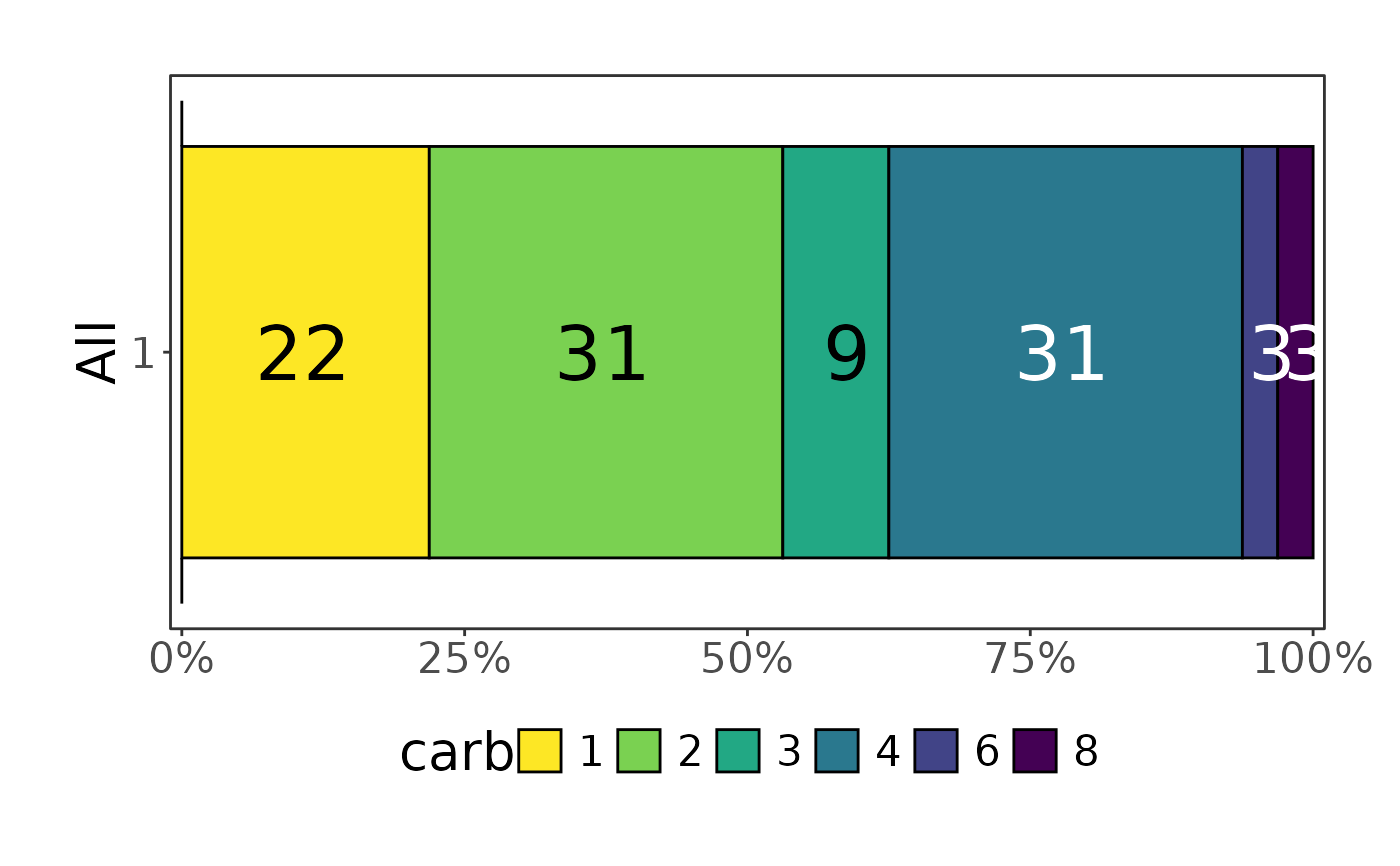
 mtcars |> plot_hbars(x = "carb", y = NULL)
#> Scale for fill is already present.
#> Adding another scale for fill, which will replace the existing scale.
mtcars |> plot_hbars(x = "carb", y = NULL)
#> Scale for fill is already present.
#> Adding another scale for fill, which will replace the existing scale.
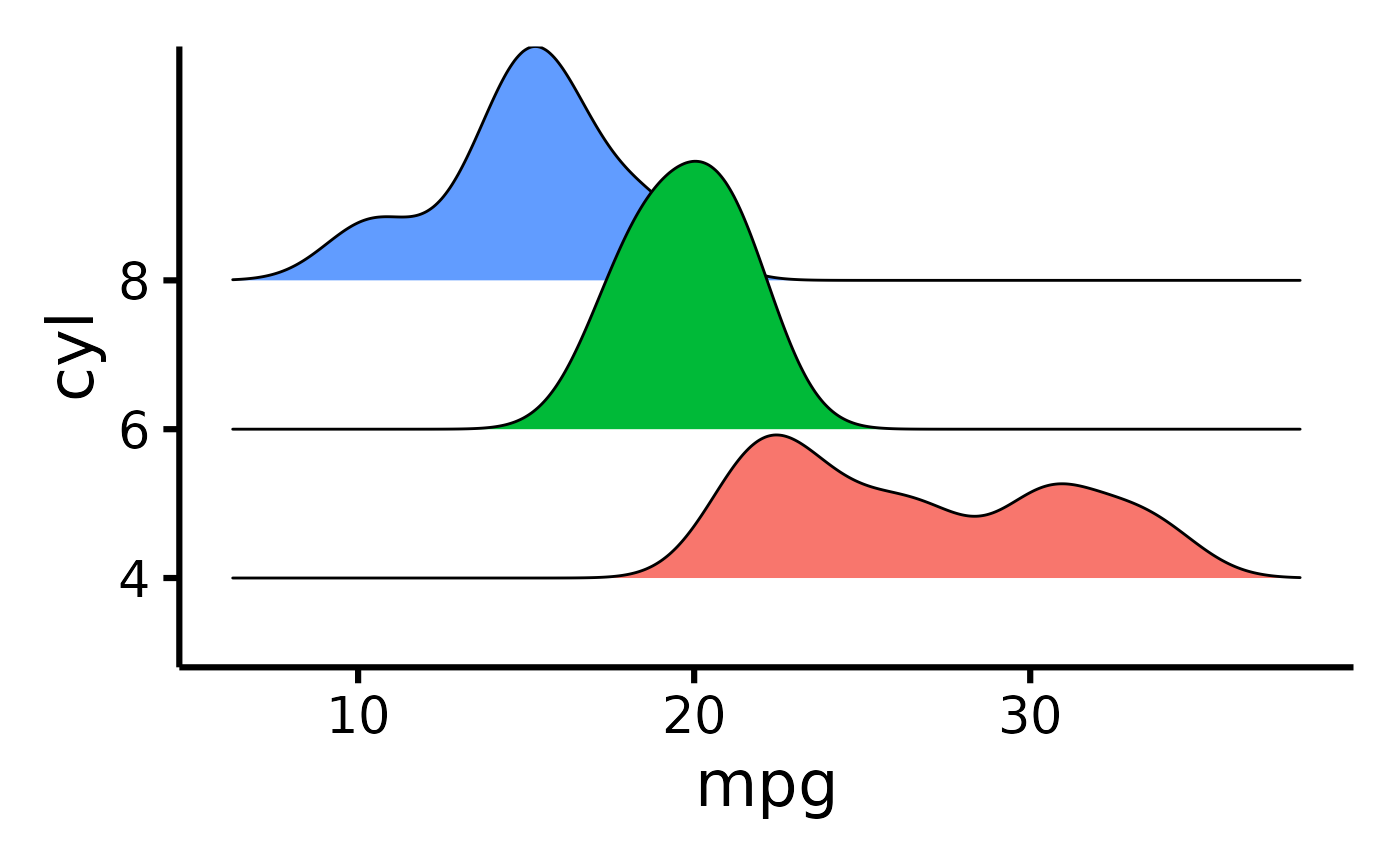
 mtcars |>
default_parsing() |>
plot_ridge(x = "mpg", y = "cyl")
#> Picking joint bandwidth of 1.38
mtcars |>
default_parsing() |>
plot_ridge(x = "mpg", y = "cyl")
#> Picking joint bandwidth of 1.38
 mtcars |> plot_ridge(x = "mpg", y = "cyl", z = "gear")
#> Picking joint bandwidth of 1.52
#> Warning: The following aesthetics were dropped during statistical transformation: y and
#> fill.
#> ℹ This can happen when ggplot fails to infer the correct grouping structure in
#> the data.
#> ℹ Did you forget to specify a `group` aesthetic or to convert a numerical
#> variable into a factor?
#> Error in ggridges::geom_density_ridges(): Problem while setting up geom.
#> ℹ Error occurred in the 1st layer.
#> Caused by error in `compute_geom_1()`:
#> ! `geom_density_ridges()` requires the following missing aesthetics: y.
ds <- data.frame(g = sample(LETTERS[1:2], 100, TRUE), first = REDCapCAST::as_factor(sample(letters[1:4], 100, TRUE)), last = sample(c(letters[1:4], NA), 100, TRUE, prob = c(rep(.23, 4), .08)))
ds |> sankey_ready("first", "last")
#> # A tibble: 19 × 7
#> first last n gx.sum gy.sum lx ly
#> <fct> <fct> <int> <int> <int> <fct> <fct>
#> 1 c c 5 28 15 "c\n(n=28)" "c\n(n=15)"
#> 2 c a 7 28 32 "c\n(n=28)" "a\n(n=32)"
#> 3 c b 6 28 26 "c\n(n=28)" "b\n(n=26)"
#> 4 c d 6 28 17 "c\n(n=28)" "d\n(n=17)"
#> 5 c NA 4 28 10 "c\n(n=28)" NA
#> 6 a c 6 24 15 "a\n(n=24)" "c\n(n=15)"
#> 7 a a 9 24 32 "a\n(n=24)" "a\n(n=32)"
#> 8 a b 6 24 26 "a\n(n=24)" "b\n(n=26)"
#> 9 a d 1 24 17 "a\n(n=24)" "d\n(n=17)"
#> 10 a NA 2 24 10 "a\n(n=24)" NA
#> 11 b c 4 20 15 "b\n(n=20)" "c\n(n=15)"
#> 12 b a 8 20 32 "b\n(n=20)" "a\n(n=32)"
#> 13 b b 5 20 26 "b\n(n=20)" "b\n(n=26)"
#> 14 b d 2 20 17 "b\n(n=20)" "d\n(n=17)"
#> 15 b NA 1 20 10 "b\n(n=20)" NA
#> 16 d a 8 28 32 "d\n(n=28)" "a\n(n=32)"
#> 17 d b 9 28 26 "d\n(n=28)" "b\n(n=26)"
#> 18 d d 8 28 17 "d\n(n=28)" "d\n(n=17)"
#> 19 d NA 3 28 10 "d\n(n=28)" NA
ds |> sankey_ready("first", "last", numbers = "percentage")
#> # A tibble: 19 × 7
#> first last n gx.sum gy.sum lx ly
#> <fct> <fct> <int> <int> <int> <fct> <fct>
#> 1 c c 5 28 15 "c\n(28%)" "c\n(15%)"
#> 2 c a 7 28 32 "c\n(28%)" "a\n(32%)"
#> 3 c b 6 28 26 "c\n(28%)" "b\n(26%)"
#> 4 c d 6 28 17 "c\n(28%)" "d\n(17%)"
#> 5 c NA 4 28 10 "c\n(28%)" NA
#> 6 a c 6 24 15 "a\n(24%)" "c\n(15%)"
#> 7 a a 9 24 32 "a\n(24%)" "a\n(32%)"
#> 8 a b 6 24 26 "a\n(24%)" "b\n(26%)"
#> 9 a d 1 24 17 "a\n(24%)" "d\n(17%)"
#> 10 a NA 2 24 10 "a\n(24%)" NA
#> 11 b c 4 20 15 "b\n(20%)" "c\n(15%)"
#> 12 b a 8 20 32 "b\n(20%)" "a\n(32%)"
#> 13 b b 5 20 26 "b\n(20%)" "b\n(26%)"
#> 14 b d 2 20 17 "b\n(20%)" "d\n(17%)"
#> 15 b NA 1 20 10 "b\n(20%)" NA
#> 16 d a 8 28 32 "d\n(28%)" "a\n(32%)"
#> 17 d b 9 28 26 "d\n(28%)" "b\n(26%)"
#> 18 d d 8 28 17 "d\n(28%)" "d\n(17%)"
#> 19 d NA 3 28 10 "d\n(28%)" NA
data.frame(
g = sample(LETTERS[1:2], 100, TRUE),
first = REDCapCAST::as_factor(sample(letters[1:4], 100, TRUE)),
last = sample(c(TRUE, FALSE, FALSE), 100, TRUE)
) |>
sankey_ready("first", "last")
#> # A tibble: 8 × 7
#> first last n gx.sum gy.sum lx ly
#> <fct> <fct> <int> <int> <int> <fct> <fct>
#> 1 a FALSE 21 29 61 "a\n(n=29)" "FALSE\n(n=61)"
#> 2 a TRUE 8 29 39 "a\n(n=29)" "TRUE\n(n=39)"
#> 3 d FALSE 12 22 61 "d\n(n=22)" "FALSE\n(n=61)"
#> 4 d TRUE 10 22 39 "d\n(n=22)" "TRUE\n(n=39)"
#> 5 b FALSE 13 26 61 "b\n(n=26)" "FALSE\n(n=61)"
#> 6 b TRUE 13 26 39 "b\n(n=26)" "TRUE\n(n=39)"
#> 7 c FALSE 15 23 61 "c\n(n=23)" "FALSE\n(n=61)"
#> 8 c TRUE 8 23 39 "c\n(n=23)" "TRUE\n(n=39)"
ds <- data.frame(g = sample(LETTERS[1:2], 100, TRUE), first = REDCapCAST::as_factor(sample(letters[1:4], 100, TRUE)), last = REDCapCAST::as_factor(sample(letters[1:4], 100, TRUE)))
ds |> plot_sankey("first", "last")
#> Loading required package: ggplot2
mtcars |> plot_ridge(x = "mpg", y = "cyl", z = "gear")
#> Picking joint bandwidth of 1.52
#> Warning: The following aesthetics were dropped during statistical transformation: y and
#> fill.
#> ℹ This can happen when ggplot fails to infer the correct grouping structure in
#> the data.
#> ℹ Did you forget to specify a `group` aesthetic or to convert a numerical
#> variable into a factor?
#> Error in ggridges::geom_density_ridges(): Problem while setting up geom.
#> ℹ Error occurred in the 1st layer.
#> Caused by error in `compute_geom_1()`:
#> ! `geom_density_ridges()` requires the following missing aesthetics: y.
ds <- data.frame(g = sample(LETTERS[1:2], 100, TRUE), first = REDCapCAST::as_factor(sample(letters[1:4], 100, TRUE)), last = sample(c(letters[1:4], NA), 100, TRUE, prob = c(rep(.23, 4), .08)))
ds |> sankey_ready("first", "last")
#> # A tibble: 19 × 7
#> first last n gx.sum gy.sum lx ly
#> <fct> <fct> <int> <int> <int> <fct> <fct>
#> 1 c c 5 28 15 "c\n(n=28)" "c\n(n=15)"
#> 2 c a 7 28 32 "c\n(n=28)" "a\n(n=32)"
#> 3 c b 6 28 26 "c\n(n=28)" "b\n(n=26)"
#> 4 c d 6 28 17 "c\n(n=28)" "d\n(n=17)"
#> 5 c NA 4 28 10 "c\n(n=28)" NA
#> 6 a c 6 24 15 "a\n(n=24)" "c\n(n=15)"
#> 7 a a 9 24 32 "a\n(n=24)" "a\n(n=32)"
#> 8 a b 6 24 26 "a\n(n=24)" "b\n(n=26)"
#> 9 a d 1 24 17 "a\n(n=24)" "d\n(n=17)"
#> 10 a NA 2 24 10 "a\n(n=24)" NA
#> 11 b c 4 20 15 "b\n(n=20)" "c\n(n=15)"
#> 12 b a 8 20 32 "b\n(n=20)" "a\n(n=32)"
#> 13 b b 5 20 26 "b\n(n=20)" "b\n(n=26)"
#> 14 b d 2 20 17 "b\n(n=20)" "d\n(n=17)"
#> 15 b NA 1 20 10 "b\n(n=20)" NA
#> 16 d a 8 28 32 "d\n(n=28)" "a\n(n=32)"
#> 17 d b 9 28 26 "d\n(n=28)" "b\n(n=26)"
#> 18 d d 8 28 17 "d\n(n=28)" "d\n(n=17)"
#> 19 d NA 3 28 10 "d\n(n=28)" NA
ds |> sankey_ready("first", "last", numbers = "percentage")
#> # A tibble: 19 × 7
#> first last n gx.sum gy.sum lx ly
#> <fct> <fct> <int> <int> <int> <fct> <fct>
#> 1 c c 5 28 15 "c\n(28%)" "c\n(15%)"
#> 2 c a 7 28 32 "c\n(28%)" "a\n(32%)"
#> 3 c b 6 28 26 "c\n(28%)" "b\n(26%)"
#> 4 c d 6 28 17 "c\n(28%)" "d\n(17%)"
#> 5 c NA 4 28 10 "c\n(28%)" NA
#> 6 a c 6 24 15 "a\n(24%)" "c\n(15%)"
#> 7 a a 9 24 32 "a\n(24%)" "a\n(32%)"
#> 8 a b 6 24 26 "a\n(24%)" "b\n(26%)"
#> 9 a d 1 24 17 "a\n(24%)" "d\n(17%)"
#> 10 a NA 2 24 10 "a\n(24%)" NA
#> 11 b c 4 20 15 "b\n(20%)" "c\n(15%)"
#> 12 b a 8 20 32 "b\n(20%)" "a\n(32%)"
#> 13 b b 5 20 26 "b\n(20%)" "b\n(26%)"
#> 14 b d 2 20 17 "b\n(20%)" "d\n(17%)"
#> 15 b NA 1 20 10 "b\n(20%)" NA
#> 16 d a 8 28 32 "d\n(28%)" "a\n(32%)"
#> 17 d b 9 28 26 "d\n(28%)" "b\n(26%)"
#> 18 d d 8 28 17 "d\n(28%)" "d\n(17%)"
#> 19 d NA 3 28 10 "d\n(28%)" NA
data.frame(
g = sample(LETTERS[1:2], 100, TRUE),
first = REDCapCAST::as_factor(sample(letters[1:4], 100, TRUE)),
last = sample(c(TRUE, FALSE, FALSE), 100, TRUE)
) |>
sankey_ready("first", "last")
#> # A tibble: 8 × 7
#> first last n gx.sum gy.sum lx ly
#> <fct> <fct> <int> <int> <int> <fct> <fct>
#> 1 a FALSE 21 29 61 "a\n(n=29)" "FALSE\n(n=61)"
#> 2 a TRUE 8 29 39 "a\n(n=29)" "TRUE\n(n=39)"
#> 3 d FALSE 12 22 61 "d\n(n=22)" "FALSE\n(n=61)"
#> 4 d TRUE 10 22 39 "d\n(n=22)" "TRUE\n(n=39)"
#> 5 b FALSE 13 26 61 "b\n(n=26)" "FALSE\n(n=61)"
#> 6 b TRUE 13 26 39 "b\n(n=26)" "TRUE\n(n=39)"
#> 7 c FALSE 15 23 61 "c\n(n=23)" "FALSE\n(n=61)"
#> 8 c TRUE 8 23 39 "c\n(n=23)" "TRUE\n(n=39)"
ds <- data.frame(g = sample(LETTERS[1:2], 100, TRUE), first = REDCapCAST::as_factor(sample(letters[1:4], 100, TRUE)), last = REDCapCAST::as_factor(sample(letters[1:4], 100, TRUE)))
ds |> plot_sankey("first", "last")
#> Loading required package: ggplot2
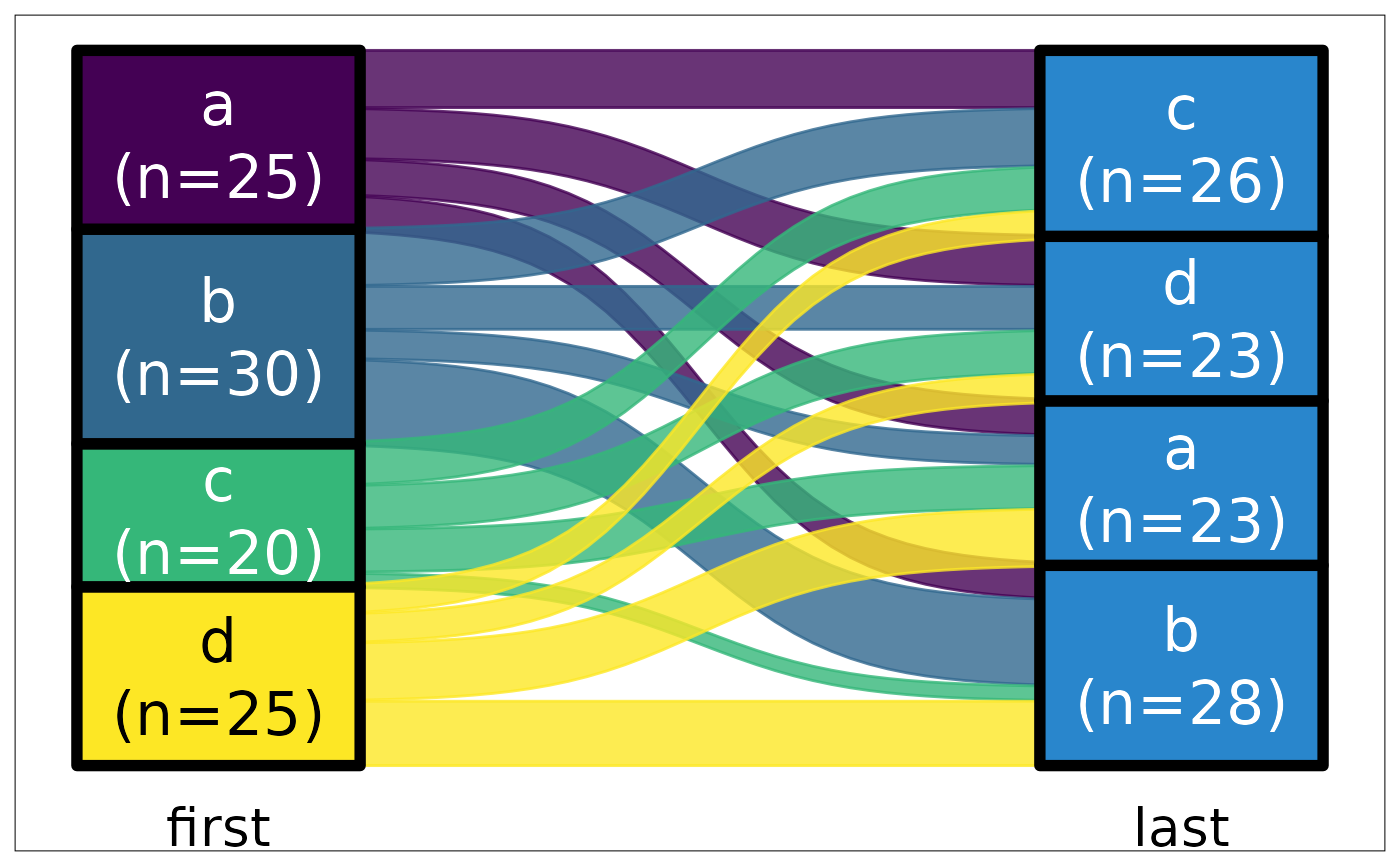 ds |> plot_sankey("first", "last", color.group = "y")
ds |> plot_sankey("first", "last", color.group = "y")
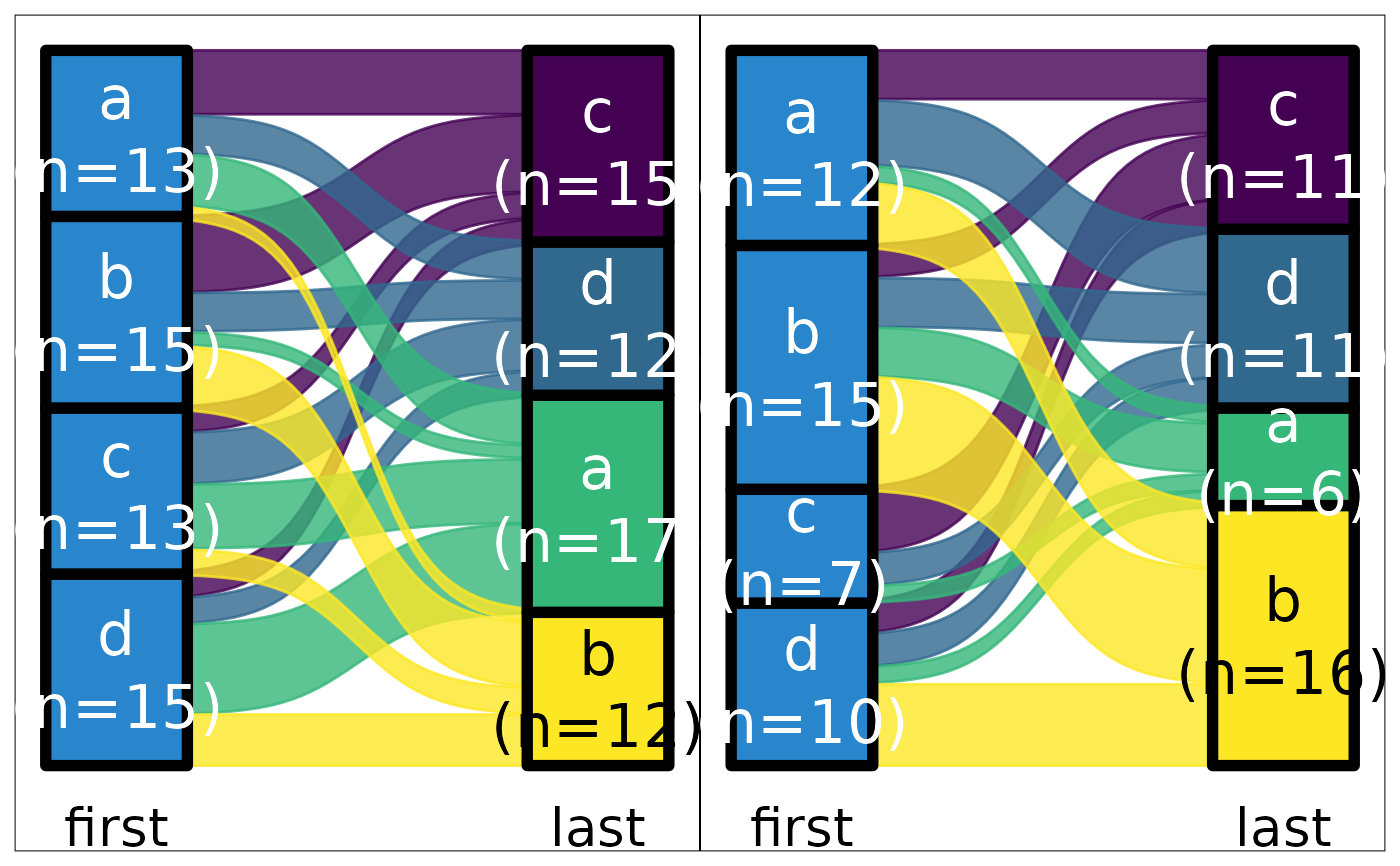
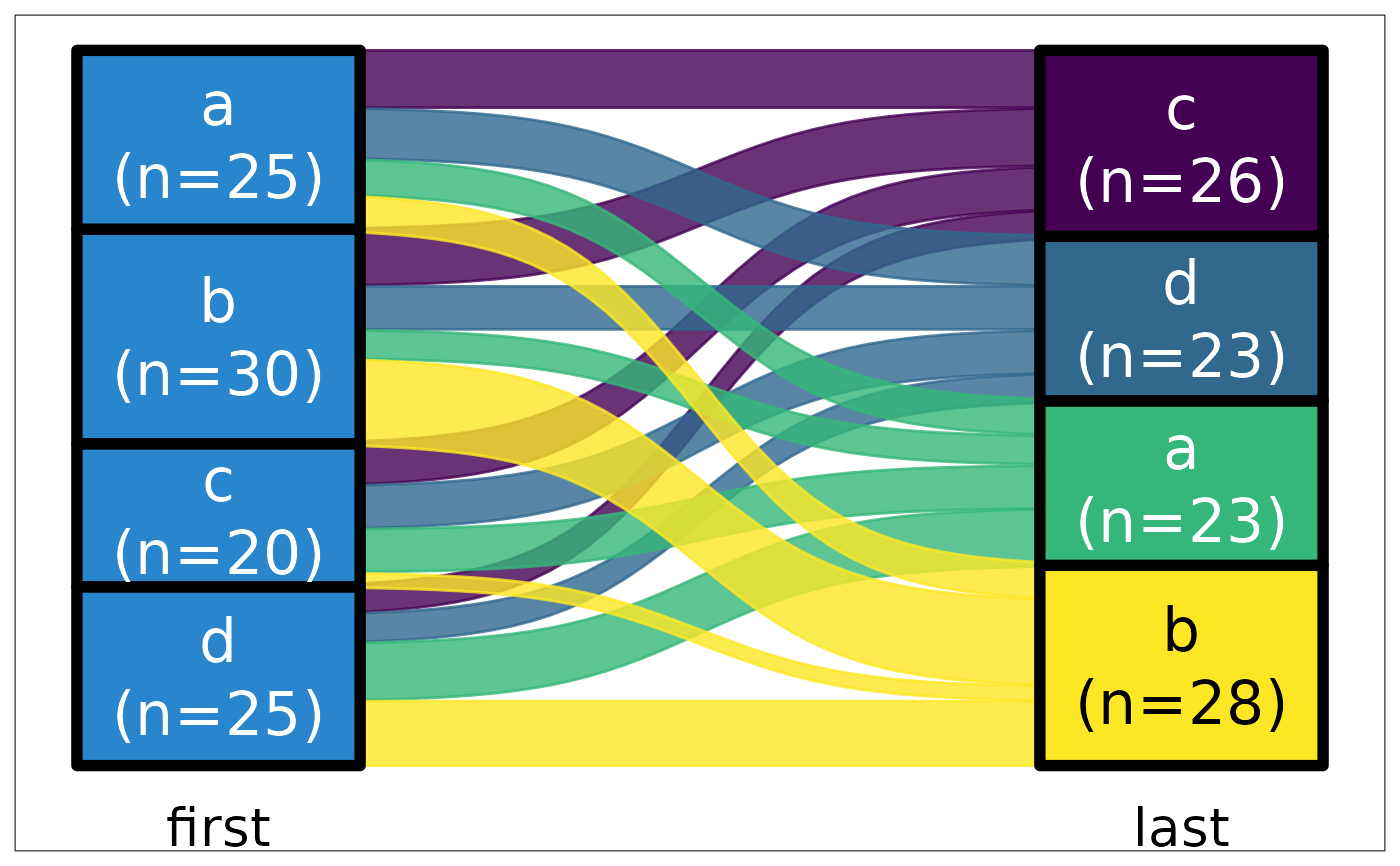 ds |> plot_sankey("first", "last", z = "g", color.group = "y")
ds |> plot_sankey("first", "last", z = "g", color.group = "y")
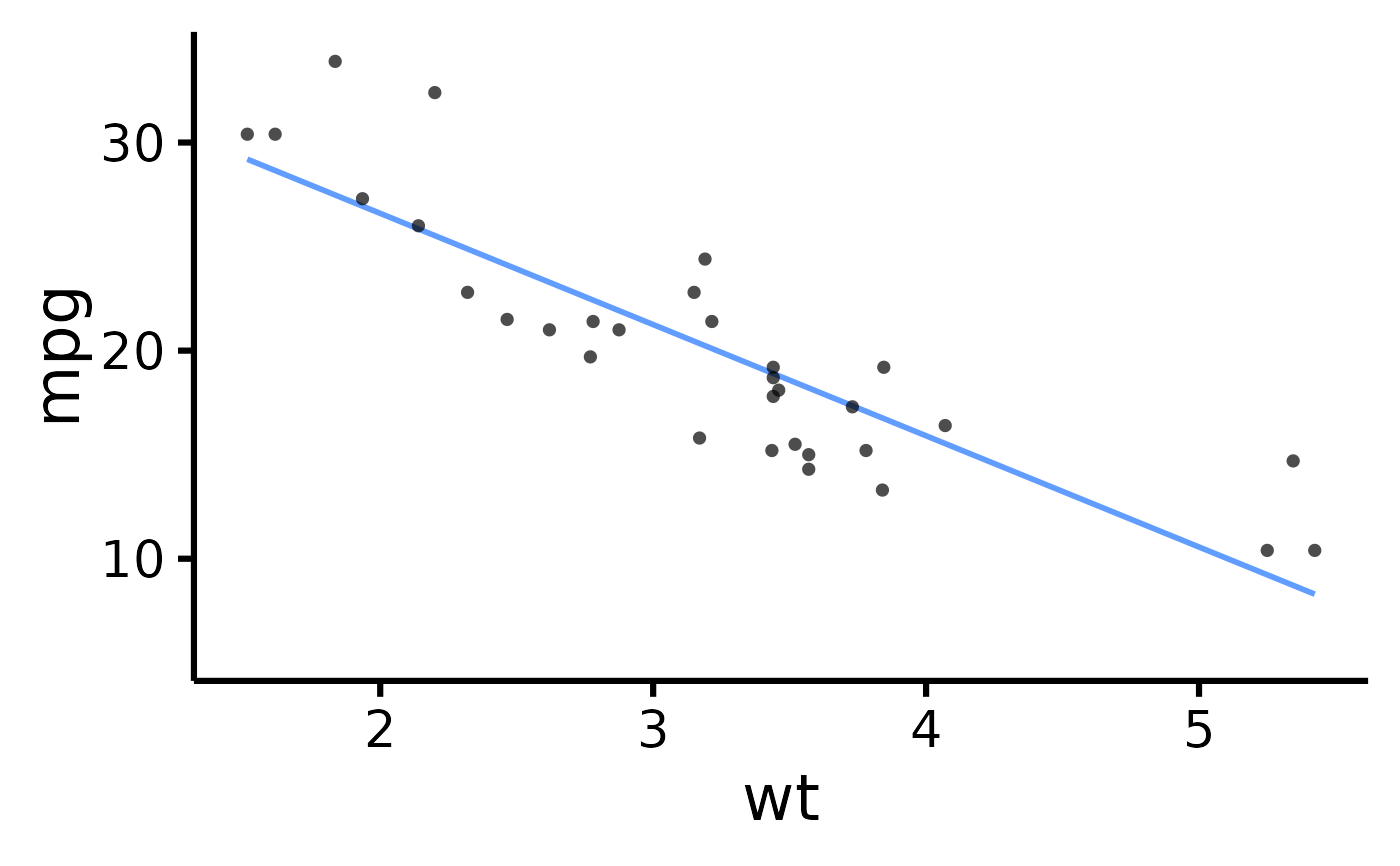
 mtcars |> plot_scatter(x = "mpg", y = "wt")
mtcars |> plot_scatter(x = "mpg", y = "wt")
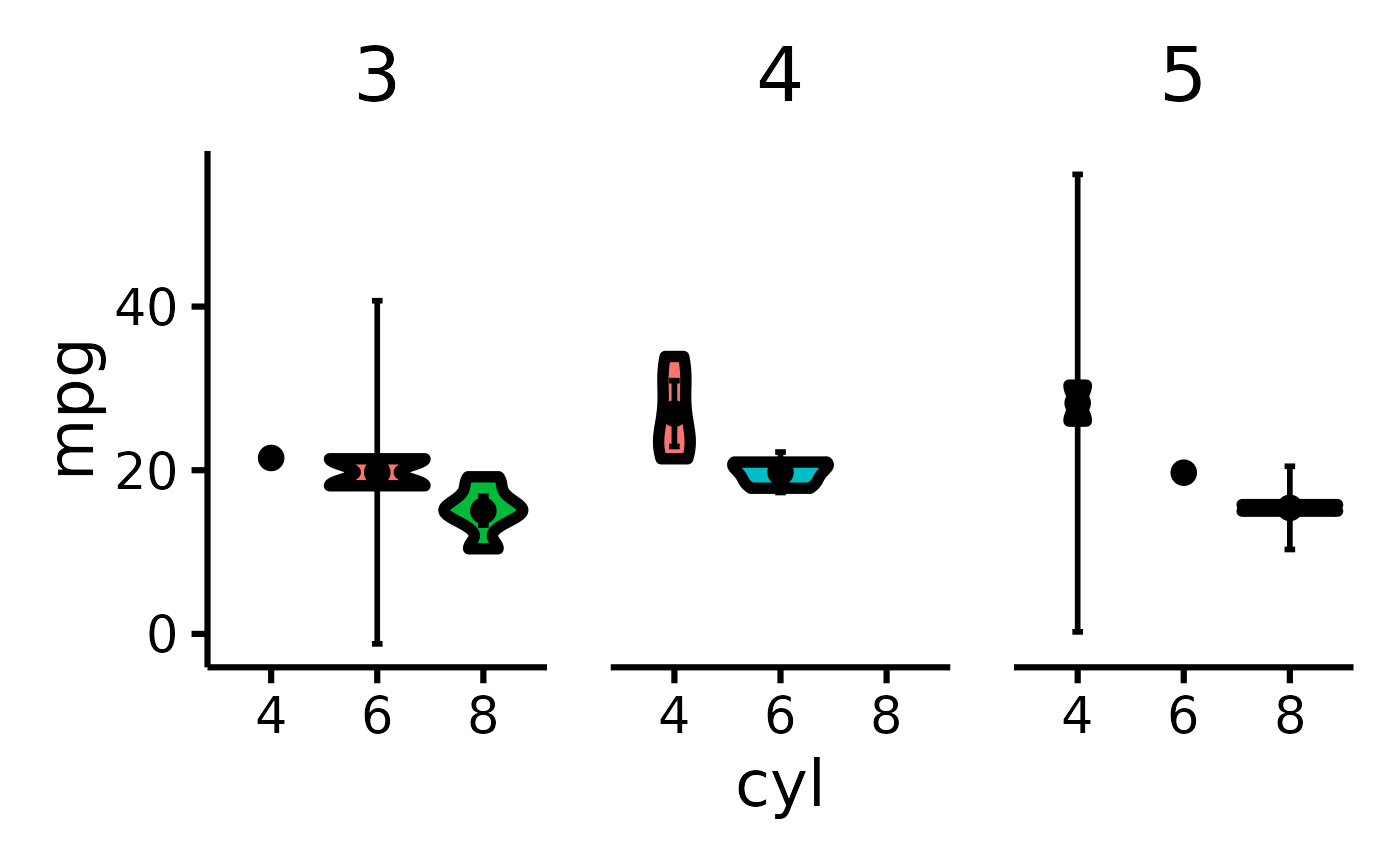
 mtcars |> plot_violin(x = "mpg", y = "cyl", z = "gear")
#> Warning: There was 1 warning in `summarize()`.
#> ℹ In argument: `V1 = .fun(as.data.frame(pick(everything())), var)`.
#> ℹ In group 1: `cyl = 4`.
#> Caused by warning in `stats::qt()`:
#> ! NaNs produced
#> Warning: There was 1 warning in `summarize()`.
#> ℹ In argument: `V1 = .fun(as.data.frame(pick(everything())), var)`.
#> ℹ In group 1: `cyl = 4`.
#> Caused by warning in `stats::qt()`:
#> ! NaNs produced
#> Warning: There was 1 warning in `summarize()`.
#> ℹ In argument: `V1 = .fun(as.data.frame(pick(everything())), var)`.
#> ℹ In group 2: `cyl = 6`.
#> Caused by warning in `stats::qt()`:
#> ! NaNs produced
#> Warning: There was 1 warning in `summarize()`.
#> ℹ In argument: `V1 = .fun(as.data.frame(pick(everything())), var)`.
#> ℹ In group 2: `cyl = 6`.
#> Caused by warning in `stats::qt()`:
#> ! NaNs produced
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
mtcars |> plot_violin(x = "mpg", y = "cyl", z = "gear")
#> Warning: There was 1 warning in `summarize()`.
#> ℹ In argument: `V1 = .fun(as.data.frame(pick(everything())), var)`.
#> ℹ In group 1: `cyl = 4`.
#> Caused by warning in `stats::qt()`:
#> ! NaNs produced
#> Warning: There was 1 warning in `summarize()`.
#> ℹ In argument: `V1 = .fun(as.data.frame(pick(everything())), var)`.
#> ℹ In group 1: `cyl = 4`.
#> Caused by warning in `stats::qt()`:
#> ! NaNs produced
#> Warning: There was 1 warning in `summarize()`.
#> ℹ In argument: `V1 = .fun(as.data.frame(pick(everything())), var)`.
#> ℹ In group 2: `cyl = 6`.
#> Caused by warning in `stats::qt()`:
#> ! NaNs produced
#> Warning: There was 1 warning in `summarize()`.
#> ℹ In argument: `V1 = .fun(as.data.frame(pick(everything())), var)`.
#> ℹ In group 2: `cyl = 6`.
#> Caused by warning in `stats::qt()`:
#> ! NaNs produced
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.